The Key to Designing a Kinase Screening Project
The key to designing any scientific study is to carefully define the question you are asking – and designing a kinase inhibitor screening project is no different. Broad selectivity kinase inhibitor profiling – such as DiscoverX’s KINOMEscan® platform – rapidly annotates a compound’s selectivity and potency across the kinome and can serve as a vital tool across a wide spectrum of the drug discovery process.
Screening Concentration – The Key to a Successful Screen
Often the most important decision made in designing a large kinase screening project is selecting a screening concentration – where the ideal concentration will largely depend on the goal of the project.
Historically, many kinase inhibitor screens have been performed at 10 µM. This screening concentration is ideal if the goal of your project is to:
- Make a direct comparison of the selectivity profile of your molecule to a known inhibitor with published data at 10 µM
- Show exquisite selectivity of your kinase inhibitor – even at a high concentration – across the kinome (think lapatinib)
- Demonstrate that your molecule – an inhibitor of another target class – has no cross-reactivity with any kinase targets
However, for many projects, a screen at 10 µM may be too high and will simply lead to profiles where too many interactions are picked up, many of which are not important to the researcher, complicating both data interpretation and potential follow-up.
How to Identify Most Potent Compounds
Utilizing a screening concentration of 1 µM or 0.1 µM will identify the most potent interactions from a screening deck, while weeding out lower affinity interactions that may not be of interest. In our experience, we have found that screening at multiple concentrations, such as 1 µM and 0.1 µM, vastly enriches the data set as it gives an assessment of overall potency and selectivity. The benefits of screening at multiple concentrations include:
- Achieve insight into “relative selectivity” by comparing selectivity profiles at both doses
- Better estimate compound affinity
- Identify potential false positives by carefully monitoring of dose responses
In summary, screening at multiple concentrations helps in both interpretation of the data set as well as allowing researchers to design effective and efficient follow-up studies.
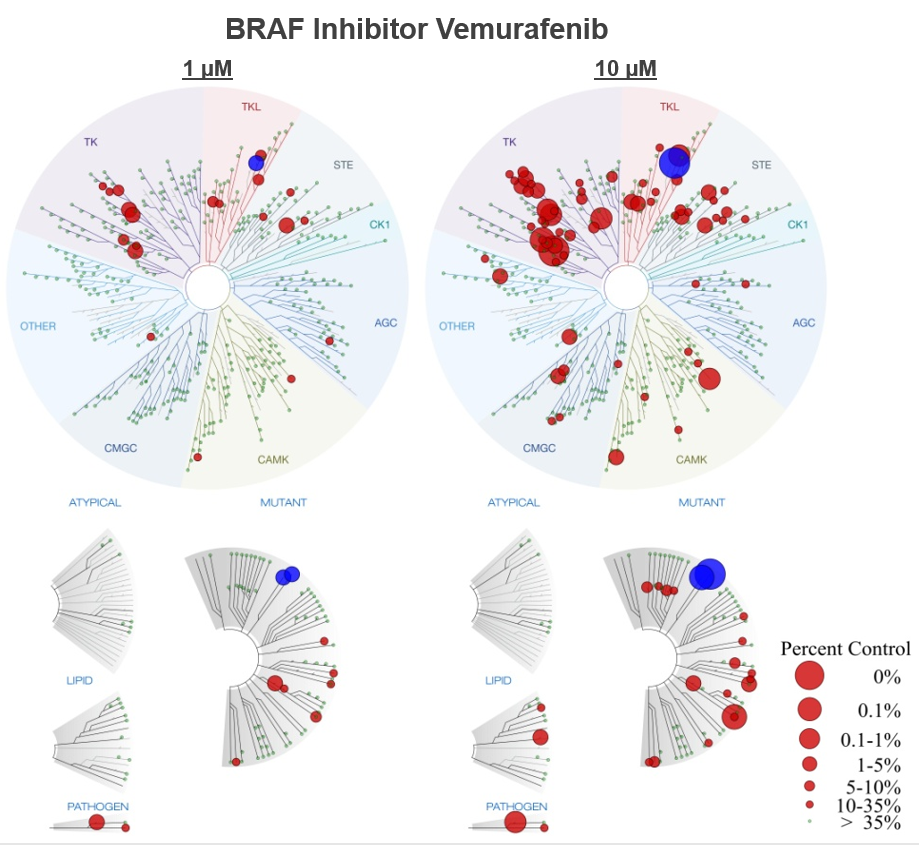
This figure demonstrations the utility of multiple concentration screens. Low affinity interactions are not detected in the 1 µM screen, helping to focus follow-up efforts.
How to Approach Homogeneous vs. Heterogeneous Assays
Understanding the screening platform being utilized for a kinase screening project is an important factor that will greatly influence the choice of an ideal screening concentration.
Homogeneous Assays
In homogeneous assays, interactions with affinities that are equal to the screening concentration are often robustly detected. Therefore, if one is interested in determining interactions 1 µM and below, then choosing a screening concentration of 1 µM would be ideal.
Heterogeneous Assays
In heterogeneous assays – such as the one employed by the KINOMEscan platform – interactions are more robustly determined when the screening concentration is greater than the affinity for the targets, typically 3 – 10x higher.
When choosing a screening concentration where a heterogeneous assay may be utilized, first carefully define an affinity threshold for which you would be interested, then choose a concentration ~3 – 10x higher.
- Example: Interested in interactions with affinities ≤100 nM? Perform the screen at 1 µM.
One of the benefits of running a heterogeneous assay includes eliminating compound interference with the assay readout, resulting in fewer false positives – thus simplifying data interpretation, and maximizing the value of potential follow-up studies.
Additionally, these assays are also well suited for screening fragment-based collections or novel chemical matter where expected affinities are low, thus screening concentrations are relatively high (>100 µM). Elimination of readout-specific false positives caused by compound interference is key for interpreting results from fragment-based screens and reducing the scope of a potentially costly follow-up study.
Start Your Kinase Screening Project Today
Whether you are trying to identify novel kinase inhibitors, demonstrate selectivity of your current compound, or identify the most potent compounds in your library, DiscoverX’s KINOMEscan Kinase Profiling & Screening Service can help you design and perform a highly informative and valuable compound screen to suit your unique drug discovery needs. Learn more and request a no obligation quote by visiting our website.
