SPRINTer Platform: Detection of Endogenous Protein Turnover by Targeted Degraders
A novel and growing modality in drug discovery has been the application of Targeted Protein Degradation (TPD). TPD involves the use of degraders (bi-functional molecules or molecular glues) to enable the degradation of intracellular target proteins expressed overabundantly (like oncoproteins in cancers) via the ubiquitin-proteasome or other cellular degradation pathways. The proteolysis-targeting chimeras (PROTACs) is among the best of several known protein degraders. They exemplify how this new class of therapeutic agents exploits cellular quality control machinery to selectively degrade target proteins, thereby overcoming traditional small molecule inhibitors development strategies.
| Advantages of Targeted Protein Degraders Over Small Molecule Inhibitors | ||
|---|---|---|
|
|
|
Assays for Protein Degradation
Assays such as western blotting that have traditionally been used to determine the potency and efficacy of protein degradation suffer from being low throughput, a high level of hands-on time, and qualitative. An ideal assay platform should be quantitative, cell-based, and compatible with the disease cell model expressing physiologically relevant levels of the target protein and E3 ligase (the enzyme involved in ubiquitin-mediated destruction of target protein). The platform should also be robust, reproducible, and sensitive to detect low protein turnovers, and amenable to high throughput screening.
The SPRINTer™ (Sensing Protein Internal Turnover) protein turnover assay platform from Eurofins DiscoverX enables sensitive detection of endogenous protein changes, such as degradation induced by PROTACs. Using both CRISPR/Cas9 and Enzyme Fragment Complementation (EFC) (discoverx.com/EFC) technologies, these assays generate a chemiluminescent readout that quantitatively reflects endogenous protein turnover. The EFC technology by Eurofins DiscoverX is comprised of two inactive β-galactosidase (β-gal) fragments; a smaller enzyme donor (ED) fragment and a larger enzyme acceptor (EA) fragment. Using CRISPR/Cas9 approach, the ED tag was incorporated into the endogenous locus of the target gene in a physiologically relevant cell model to generate an ED-tagged target protein. The addition of the EA fragment along with detection reagents enables complementation of the ED and EA fragments resulting in an active β-gal enzyme that hydrolyzes a substrate and generates a chemiluminescent signal. In the presence of degraders such as PROTAC, the ED-tagged protein is degraded, and hence EA fails to complement with the ED. This results in a decreased signal, as shown in Figure 1.
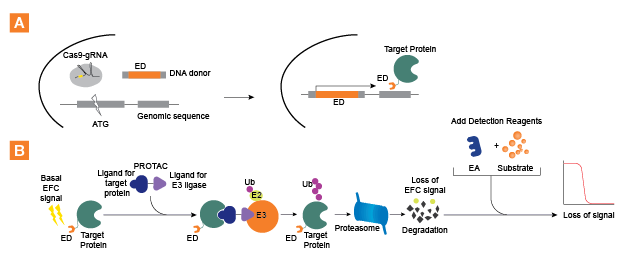
Figure 1. SPRINTer platform assay principles. A. A schematic illustration of CRISPR mediated knock-in of ED tag (one of the two recombinant β-gal fragments of the EFC technology) into a target locus to create an ED-fusion protein for EFC assay. Cas9, guide RNA, and a donor DNA encoding the ED fragment are transfected into cells. Cell clones with robust EFC signals are isolated and optimized for protein turnover assay. B. A graphic representation of EFC detection of target protein degradation induced by PROTACs (trademark of Arvinas). The endogenous ED-target fusion protein (which produces a moderate to high basal EFC signal) is brought into close proximity to the protein degradation machinery by a PROTAC, a bi-functional molecule that bridges the target protein and a specific endogenous E3 ligase. This proximity leads to the subsequent degradation of the target protein resulting in loss of EFC signal. Hence, the kinetics of protein turnover induced by PROTACs can be quantified using EFC, and is amenable to medium to high throughput screening modalities.
As a proof-of-concept, Eurofins DiscoverX applied this assay platform to test a panel of PROTACs targeting bromodomain-containing protein 4 (BRD4), and its downstream target, c-Myc, in blood cancer cell models. BRD4 inhibitors, BRD4-PROTACs, and control molecules were treated with BRD4 SPRINTer cells and evaluated by the EFC assay. As a comparison, a commonly used phenotypic endpoint assay of cell proliferation was included. The results show that SPRINTer BRD4 and c-Myc cells displayed high sensitivity and more rapid kinetics of protein degradation than CellTiter Glo® proliferation assay. The potency rank ordering of these PROTACs observed using SPRINTer BRD4 and c-Myc cells is consistent with previous reports. The c-Myc SPRINTer cell line provides an opportunity for screening molecules that directly modulate c-Myc stability.
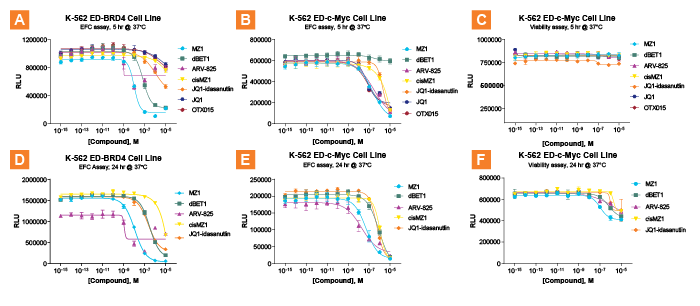
Figure 2. Time course of endogenous protein degradation. SPRINTer K-562 ED-BRD4 cells were treated with BRD4 inhibitors (JQ1 and OTX015), BRD4 PROTACs (MZ1, dBET1, JQ1-idasanutlin and ARV-825), or control molecule (cisMZ1) for 5 hr (A.) and 24 hr (D.) and evaluated by the EFC assay. The degradation of c-Myc protein induced by the same treatments of molecules using the K-562 ED-c-Myc cell line are shown in B. and E. A set of sister plates with identical treatments was also set up for cell viability assay using CellTiter Glo (Promega) (C. and F.). Results indicate the EFC assay quantifies PROTAC-mediated BRD4 protein turnover as early as 5 hours (A.), allowing rank ordering of PROTAC potency and efficacy (ARV825 > MZ1 >> dBET1 > JQ1-idasanutlin). Rank order changes slightly with longer incubations. As previously reported, the two small molecule BRD4 inhibitors, JQ1 and OTX015, are much less efficacious at mediating BRD4 degradation than the PROTACs (A. and D.). In contrast, BRD4-targeting PROTACs mediated degradation of ED-c-Myc with slightly slower kinetics, produces more distinct profiles after 24 hr incubation (E.). Nevertheless, the K-562 ED-c-Myc cell line provides equivalent rank order to that of K-562 ED-BRD4.
A wide range of applications for SPRINTer cell lines is also established, allowing the identification of degraders for BTK tyrosine kinase, inhibitors for the MDM2/p53 E3 ligase complex, and molecular glues for cereblon. In addition, SPRINTer cell lines have been adapted to the InCELL Pulse™ Compound-Target Engagement assay platform to allow for determining compound cell membrane penetration and measuring direct binding between compounds and the specific target protein. As proof-of-concept, the SPRINTer BRD4 and BTK cell lines were adapted to the InCELL Pulse assay, and rank order of the binding affinities of inhibitors were determined. This data is shown in the scientific poster (link below).
Conclusions
A simple and homogeneous assay format to quantify the modulation of endogenous protein levels in response to therapeutic agents is available as SPRINTer protein turnover assays. Moreover, the combined use of CRISPR/cas9 technology with the EFC system brings out direct and rapid quantitation of drug-induced changes in endogenous target protein levels. The high sensitivity of the SPRINTer cell-based assay platform allows for the detection of target protein turnover induced by PROTACs with more rapid kinetics than phenotypic endpoint assays, such as cell proliferation.
Resources
- Learn more about the SPRINTer targeted protein degradation assay platform at (discoverx.com/protein-degradation)
- Knowledge-Based Video: Detection of Protein Turnover Induced by Targeted Degraders
- Blog: Rapidly Identify Therapeutics that Modulate Protein Levels Induced by Protein Degraders such as PROTACs
- Webinar: Protein Degradation (PROTACs/CRISPR) Assays
- Poster: SPRINTer Platform: Detection of Endogenous Protein Turnover and Target Engagement of Targeted Degraders
