Rapidly Identify Therapeutics that Modulate Protein Levels Induced by Protein Degraders such as PROTACs
As newer, cutting-edge therapeutic technologies emerge, an increasing need for rapid and reliable screening tools follows. Recent advances in the field of protein degrader therapeutics—such as PROTAC® and molecular glues—call for analytical methods that accurately measure the on-target activity of a therapeutic. This blog briefly discusses how PROTACs influence endogenous protein turnover, and how this change in protein levels can be accurately measured using a novel cell-based, physiologically relevant assay.
What are PROTACs?
A PROTAC—or proteolysis targeting chimera—is a heterobifunctional molecule that harnesses the cell’s natural ubiquitin-proteasome degradation system to destroy disease-causing proteins. It consists of two small molecules connected by a linker. One small molecule binds to the target protein, while the other binds to an E3 ligase. Different from conventional small molecules that typically occupy an active site of an enzyme or target, these PROTACs drive protein degradation by bringing the target protein and E3 ligase in close proximity. As a result, the target protein is polyubiquitinated and subsequently degraded, as shown in Figure 1. This approach has allowed previously ineffective small molecules to be retooled as PROTACs for next-generation therapeutics through protein degradation rather than inhibiting a target protein’s enzymatic activity. Furthermore, it has been found that relatively low concentrations of PROTACs are sufficient to facilitate protein degradation. Therefore, PROTACs are able to address certain limitations that are posed by conventional small molecules.
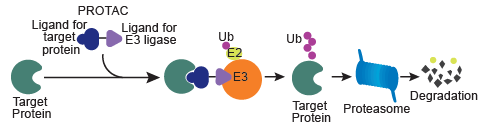
Figure 1: Principle of a PROTAC. The heterobifunctional molecules carries two ligands, one that binds to the target protein, and the other for recruiting E3 ligase. The linker that holds these ligands together helps in bringing the target protein and E3 ligase into close proximity, thus facilitating polyubiquitination and proteolysis.
Assays to Measure Endogenous Protein Turnover
The obvious question then arises: how do you confirm that a PROTAC modulates your target of interest effectively? Typically, assays used for monitoring PROTAC-induced protein turnover include Western blots, ELISAs, proliferations assays, viability assays, etc. While they relay relevant information, they are either limited by availability of robust antibodies, low-throughput formats, and/or longer assay run times. Therefore, analysis of target turnover would benefit from a more rapid, scalable, and reproducible method for measure endogenous protein turnover.
SPRINTer™ Protein Turnover Biosensor Assays provide a high throughput screening platform to identify new molecular entities that modulate endogenous protein levels for therapeutic development. Using CRISPR/Cas9 and Enzyme Fragment Complementation (EFC) technologies, these assays generate a chemiluminescent readout that quantitatively reflects PROTAC-mediated protein turnover. EFC comprises two inactive β-galactosidase (β-gal) fragments, a smaller enzyme donor (ED) fragment and a larger enzyme acceptor (EA) fragment. CRISPR/Cas 9 is used to incorporate the ED tag into the endogenous locus of the target gene in a physiologically relevant cell model. Expression of the target gene from its native promoter results in production of an ED-tagged target protein. If the tagged target protein remains intact, the ED tag complements with EA fragment, which is added along with the detection reagents. The complementation of ED and EA results in an active β-gal enzyme that hydrolyzes a substrate and generates a chemiluminescent signal. However, in the event of PROTAC-induced degradation, EA cannot complement with ED (due to loss of the ED-tagged protein), resulting in a decrease in signal indicative of protein turnover, as shown in Figure 2.
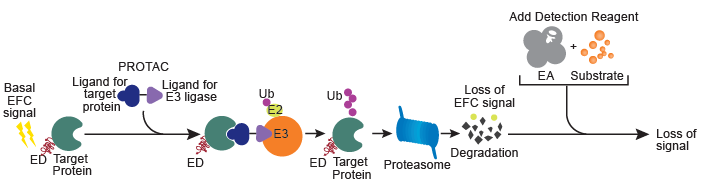
Figure 2. Measurement of PROTAC-mediated endogenous protein turnover using the SPRINTer Endogenous Protein Turnover Biosensor Cell Line. The heterobifunctional PROTAC molecule facilitates ubiquitination of the ED-tagged protein by bringing it into proximity of a specific E3 ligase. As a result, the ED-tagged protein undergoes degradation. A loss of the chemiluminescent signal is observed when the complementary enzyme fragment, EA, is added with the detection reagents.
Applications of SPRINTer Protein Turnover Biosensor Assays
As a proof-of-concept, this assay platform was designed with bromodomain-containing protein 4 (BRD4), as well as its downstream effector c-Myc, as target proteins. To achieve this, an array of PROTACs were evaluated for the degradation of BRD4 and c-Myc. Figure 3 demonstrates the rank ordering of potencies of select PROTACs and BET (bromodomain and extra-terminal motif) inhibitors (JQ1 and OTX015) in the SPRINTer K-562 cell lines. The results obtained were similar to those from previously published ELISA data1 (not shown) with minor differences that are accounted for due to different cell backgrounds used in these two different assays. Remarkably, sensitive on-target potencies were measured within 24 hours, the shortest time-frame that enabled distinguishing between therapeutic agents when compared to other assays.
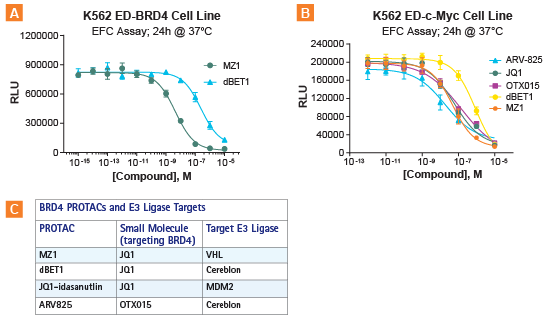
Figure 3. PROTACs were evaluated for the degradation of BRD4 and c-Myc. A. Rank ordering of PROTACs (dBET1 and MZ1) in the SPRINTer K-562 ED-BRD4 Protein Turnover Cell Line. B. Rank ordering of BET inhibitors (OTX15 and JQ1) and PROTACs (dBET1,MZ1, ARV-825) using the SPRINTer K-562 c-Myc Protein Turnover Cell Line. C. Different BRD4-targeting agents tested in this assay. The two PROTACs, MZ1 and dBET1, both contain JQ1, but engage with two different E3 ligases (VHL and cereblon, respectively). Results show that potencies were determined for these molecules after 24h treatment at 37°C, producing the following rank order: ARV825 > JQ1 = OTX015 > dBET. This rank order of potencies is similar to previously published ELISA data (not shown; from Kanak et al, (2016) PNAS 113 (26) 7124-7129) using a 22Rv1 c-myc cell line which showed ARV825 > JQ1 > OTX015 > dBET.
The SPRINTer Protein Turnover Biosensor Assays are suitable for other analyses in addition to rank ordering of drug potencies. For example, different cell lines representative of disease-relevant cell models may express varying levels of E3 ligases. As a result, efficacies of PROTACs will also vary depending on the cell background, and these differences can be captured by using different cell lines in these assays.
Conclusion
The SPRINTer Protein Turnover Biosensor Assays afford higher sensitivity and more rapid kinetics than phenotypic endpoint assays. Comparative analysis (data not shown here) demonstrates that even a short assay run time such as 5-6 hours starts to uncover the differential kinetics of endogenous protein degradation. Using blood cancer cell lines adequately expressing BRD4 and c-Myc, we have demonstrated efficient rank-ordering of various PROTAC potencies. Furthermore, these assays are not limited to the evaluation of PROTACs, but other targeted protein degraders as well. To learn more about the versatility of this assay platform, visit discoverx.com/turnover.
PROTAC® is a registered trademark of Arvinas.
SPRINTer™ is a trademark of Eurofins DiscoverX.
References
- Raina K, Lu J, Qian Y, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113(26):7124-7129. doi:10.1073/pnas.1521738113
Resources
- Blog: SPRINTer Platform: Detection of Endogenous Protein Turnover of Targeted Degraders
- Knowledge-Based Video: Detection of Protein Turnover Induced by Targeted Degraders
- Poster: Biosensor Development Using CRISPR to Quantify Endogenous Protein Expression Modulated by PROTACs
- Webinar: Protein Degradation (PROTACs/CRISPR) Assays
- Custom Assay Development
