Unlocking Orphan GPCRs with an Innovative Forward Trafficking Approach
Application of GPCR Pharmacotrafficking & Cell-Based Assays
Orphan G Protein-Coupled Receptors (oGPCRs) comprise one-third of the druggable GPCR space1. Despite their pharmacotherapeutic potential, they remain underexploited owing to difficulties in interrogating their physiological roles and signaling pathways. Further, the lack of chemical probes or antibodies needed for elucidating their roles makes oGPCRs largely untapped for pharmaceutical interventions in a variety of diseases.
To address this gap, a systematic approach to the identification of chemical probes of oGPCRs using a recently developed novel platform called the Ligand-Induced Forward Trafficking (LIFT) assay has been developed. Established to circumvent impediments to the application of automated high-throughput technologies to pursue oGPCR screening, the innovative LIFT assay design features,
- ✓ A universal assay system that is suitable for high throughput screening (HTS)
- ✓ No requirement of a priori knowledge of receptor pharmacology
- ✓ Universal positive control
- ✓ Reasonably cost-effective and integrative platform for lead ID
- ✓ Rapid ability to validate utility
Scientists at the Conrad Prebys Center for Chemical Genomics at Sanford Burnham Prebys (SBP) Medical Discovery Institute have conducted a full HTS campaign to identify chemical probes of the orphan GPCR, GPR20. They used a highly curated library of compounds in the LIFT approach thereby providing a new approach to determine compound:target engagement.
Development and overall validation of the LIFT assay platform, as well as the utility of hits emerging from this HTS campaign was conducted in collaboration with Eurofins DiscoverX using the GPCR PathHunter® Pharmacotrafficking assay, PathHunter arrestin recruitment assay, and HitHunter® cAMP assay platforms, as well as the gpcrMAX(SM) panel services.
Identification of Chemical Probes for oGPCRs
Overview of the LIFT Assay
The LIFT assay platform was developed to enable the identification of chemical probes for oGPCRs using the concept of pharmacochaperone-induced trafficking of mutant receptors. Pharmacochaperones are cell-permeable small molecules that bind to mutant receptors, stabilizes them, and enables their trafficking to the membrane2. Assays that report on receptor trafficking, such as the cell-based pharmacotrafficking assays from DiscoverX, have been valuable in the identification of target-specific ligands, and the interrogation of their mechanisms-of-action (MOA). When applied to oGPCRS, the assay consists of a target orphan receptor harboring a synthetic point mutation that causes retention of the target in the endoplasmic reticulum (ER) due to improper folding. [It is to be noted that the cellular protein synthesis machinery can accommodate point mutations usually found in conserved Cholesterol Consensus Motif (CCM) residues among many class A oGPCRs3.] Upon binding of a chemical ligand (pharmacochaperone), the mutated receptor adopts a stabilized conformation that is forward-trafficked through the golgi, to the plasma membrane, and ultimately to the early endosome. DiscoverX’s Enzyme Fragment Complementation (EFC) platform, which is the base technology of these assays, reports the forward trafficking of the receptor yielding a robust luminescent signal that can be measured. The overview of the general oGPCR LIFT lead discovery strategy is shown in Figure 1, which uses DiscoverX’s pharmacotrafficking, cAMP, and β-arrestin assays along with Eurofins Discovery services gpcrMAX panel to validate the forward trafficking approach.
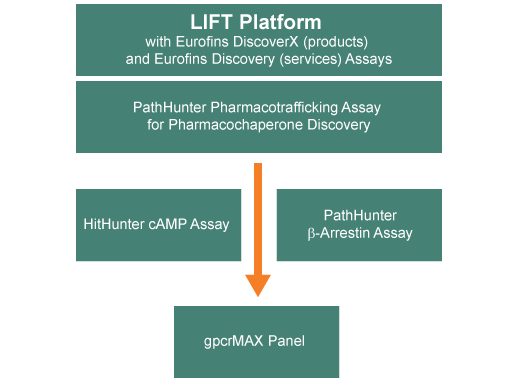
Figure 1. Flow schematic of the LIFT assay platform for ligand identification. The LIFT platform uses DiscoverX’s pharmacochaperone, cAMP, and β-arrestin assays along with Eurofins Discovery services gpcrMAX panel to validate the forward trafficking approach to identify chemical probes against oGPCRs.
As proof of concept, LIFT was applied to the human orphan GPR20, and a large compound collection (~450,000 compounds) was screened with the goal of identifying small molecule probes of GPR20. Since the approach involves mutating the target receptor, it is to be noted that the induced mutation is not related to human disease, but only as a means to retain the receptor in the ER. Hence, assays for wild-type receptor including cAMP and β-arrestin recruitment assays are required for hit validation. Given the orphan nature of GPR20, compound selectivity to GPR20 was also assessed using DiscoverX’s gpcrMAX panel assays. Overall, applying a wide array of technologies, such as second messenger, receptor internalization, and cellular impedance assays formed a comprehensive strategy for probe discovery, delineating functional cell-based assays to serve as critical tools for interrogating the MOA in drug discovery therapeutic characterization.
How Pharmacochaperone Assay Can Monitor Receptor Translocation?
Eurofins DiscoverX’s PathHunter cell-based pharmacotrafficking assays provide a simple and quantitative tool for broad pharmacological characterization and interrogation of compound function in disease processes associated with receptor protein trafficking and internalization due to protein misfolding. Featured as part of the repertoire of cell-based assays surrounding translocation and trafficking of membrane proteins, the benefits of its use over other methods are described here. These pharmacotrafficking assays provide a validated, cell-based chemiluminescent assay platform that enables identification and optimization of compounds that function as pharmacochaperones by promoting proper folding of misfolded receptor proteins and assisting in trafficking to their intended location.
Figure 2 illustrates the EFC-based PathHunter pharmacotrafficking assay in two formats wherein the binding of a chemical ligand (pharmacochaperone) to a misfolded, tagged receptor protein stabilizes the protein in a conformation that enables its trafficking through the golgi, then onward to the cell membrane, and further internalized to the endosome. Both formats involve two inactive β-galactosidase (β-gal) enzyme fragments (a small enzyme donor and a large enzyme acceptor (EA)) to come together to form an active β-gal enzyme that hydrolyzes a substrate to generate a chemiluminescent signal that can be measured on any standard luminometer. In these assays, the membrane protein (GPCR in this case) is tagged with the small enzyme donor called ProLink™ (PK) and the EA is localized to either the cell membrane or endosome. In the Membrane-EA format (Figure 2 (1)), complementation of the two β-gal enzyme fragments (EA & PK) occurs at the membrane. In contrast, in the Endosome-EA format (Figure 2 (2)), the complementation of the fragments occur in endosomes after receptor internalization. In the LIFT platform strategy, the Endosome-EA assay format has been employed for monitoring mutant receptor translocation.
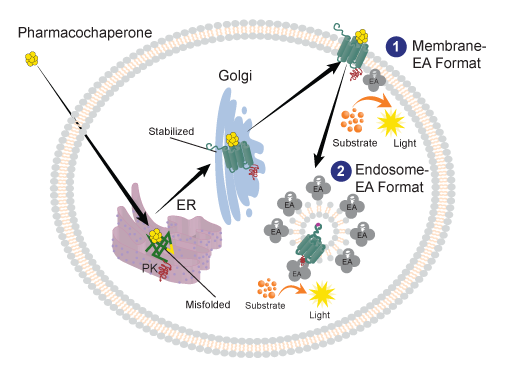
Figure 2. Pharmacochaperone Assay principle using EFC technology. 1. Membrane-EA format: Cell lines co-express the PK-tagged receptor protein that is retained in the ER (due to protein misfolding) and EA localized to the cell membrane. 2. Endosome-EA format: Cell lines co-express the PK-tagged receptor protein and the EA localized to the early endosomes. In both formats, the complementation of the two β-gal enzyme fragments (EA & PK) at either the transmembrane or endomes results in an active β-gal enzyme that hydrolyzes the substrate to generate a chemiluminescent signal.
Using the fully optimized LIFT assay on β2-Adrenergic Receptor (ADRB2), a screening of a Library of Pharmacologically Active Compounds (LOPAC) typically used for demonstrating HTS feasibility of an assay was performed as LOPAC contains several compounds with known activity at ADRB2, as well as many with no effect at this receptor. (www.sigmaaldrich.com/life-science/cell-biology/bioactive-small-molecules/lopac1280-navigator.html)4. The ADRB2 LIFT assay performed well with a Z’ factor = 0.5 and having identified all ADRB2 ligands in the LOPAC library. The complete results of this pilot study are reported in (Morfa CJ et al. 2018).
Compound Screening & Hit Confirmation
HTS screening and hit confirmation using the LIFT assay platform was employed to identify novel ligands of GPR20. The assay performed well exhibiting a Z factor = 0.5′ (4). Among the hits (> ~300 compounds), compound SBI-6630 emerged as a confirmed hit showing a robust concentration response in the GPR20 LIFT assay (EC50 = 0.067 µM) and a high degree of selectivity for GPR20 over ADRB2 (Figure 3. A.). SBI-6630 was further validated as a potent and selective GPR20 agonist using wild-type human GPR20 (hGPR20) expressed in HEK-293 cells. The ability of SBI-6630 to reduce forskolin-stimulated intracellular cAMP was measured using the HitHunter cAMP assay. The compound exhibited robust activity with a similar potency to that observed in the primary LIFT assay (EC50 = 0.150 µM). As before, SBI-6630 showed a high degree of selectivity for hGPR20 with no activity observed in cells lacking GPR20 (parental HEK-293).
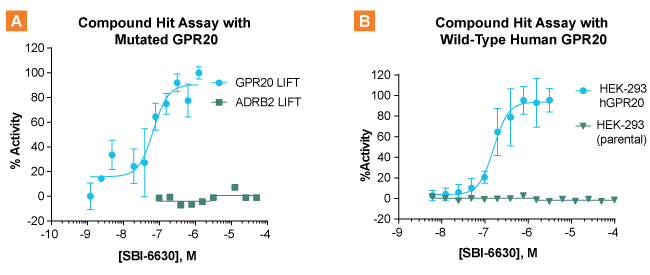
Figure 3. SBI-6630 shown as a validated hit agonist of GPR20. A. Dose-response activity of SBI-6630 against mutated GPR20 and ADBR2. B. Dose-response activity of SBI-6630 against wild-type GPR20 and parental cells lacking GPR20.
Conclusions
A novel approach that addresses critical limitations surrounding de-orphanization of GPCRs is presented as a solution. Further, employing functional downstream assays to validate hits, and confirm probe compound selectivity and receptor signaling specificity, underscores the overall value of the strategy encompassing the forward trafficking approach. Representative data shown here, in addition to data from previous work4, verifies the capability of the pharmacotrafficking assay platform to identify chemical ligands of oGPCRs using existing high-throughput screening automation and compound libraries. The implementation of DiscoverX’s EFC technology in the adoption of a forward trafficking approach not only ensures specificity, but also exerts suitability for automated screening platforms at an economical and time-sensitive standpoint, especially with the availability of target- and MOA-selective ready-to-run assays.
References
- Hopkins AL, Groom CR: The druggable genome. Nat Rev Drug Discov. 2002; 1: 727–730.
- Leidenheimer NJ, Ryder KG: Pharmacological chaperoning: a primer on mechanism and pharmacology. Pharmacol Res. 2014; 83: 10–19.
- Hanson MA, Cherezov V, Griffith MT, et al.: A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 2008; 16: 897–905.
- Morfa CJ, Bassoni D, Szabo A, McAnally D, Sharir H, Hood BL, Vasile S, Wehrman T, Lamerdin J, Smith LH. A. Pharmacochaperone-based high-throughput screening assay for the discovery of chemical probes of orphan receptors. Assay Drug Dev Technol. 2018 Oct; 16 (7): 384-396.
Resources
- Pharmacotrafficking Assays
- GPCR Products & Solutions including cAMP and calcium accumulation, β-arrestin recruitment, and internalization assays
- Orphan GPCR Target List
- EFC platform for interrogating biomolecular reactions
- Ideal Methods to Study Cellular Transport of Membrane Proteins: Translocation and Trafficking of Membrane Proteins blog
